Paper Summary
-
The methylation of protein lysine residues is an important posttranslational modification in eukaryotes. In contrast, this modification has been underexplored in prokaryotes. In this study, we analyzed the cell surface proteins of the toluene-degrading bacterium Acinetobacter sp. Tol 5 using label-free liquid chromatography‒mass spectrometry (LC‒MS) and found extensive lysine methylation in its trimeric autotransporter adhesin (TAA), AtaA. Over 130 lysine residues of AtaA, which consists of 3,630 amino acids and contains 234 lysine residues, were methylated.We identified that the outer membrane protein lysine methyltransferase (OM PKMT) of Tol 5, KmtA, specifically methylates the lysine residues of AtaA. In the KmtA-deficient mutant, most lysine methylations on AtaA were absent. The lack of KmtA also led to increased AtaA levels on the cell surface, enhanced bacterial adhesion, and slower growth, suggesting that KmtA is essential for maintaining optimal cellular adhesion. Bioinformatic analysis revealed that genes similar to KmtA are widely distributed across various pathogenic and environmental bacteria. The widespread presence of KmtA-like PKMTs throughout gram-negative bacteria suggests that lysine methylation plays a more extensive role in bacterial physiology than previously recognized.
This study was selected as an Editor's Pick in Journal of Bacteriology, published by the American Society for Microbiology. - The bacterium Acinetobacter sp. Tol 5, discovered in the Hori Laboratory, demonstrates high adhesiveness to various material surfaces, from hydrophobic plastics to hydrophilic glass and even metals, via a trimeric autotransporter adhesin (TAA) protein known as AtaA. This paper investigates the adhesive behavior of Tol 5 and other bacteria possessing TAAs on surfaces that are traditionally considered difficult for cell and biomolecule adhesion. Tol 5 was found to adhere to surfaces with low surface free energy such as polytetrafluoroethylene (Teflon), hydrophilic polymer brushes, and even atomically flat mica surfaces. Furthermore, single-cell measurements using atomic force microscopy (AFM) revealed the strong cell adhesion force of Tol 5 to these challenging surfaces. Conversely, it was found that Tol 5 hardly adheres to 2-methacryloyloxyethyl phosphorylcholine (MPC) polymer surfaces. These results contribute to the understanding and control of bacterial adhesion mechanisms mediated by TAAs, including AtaA, which are of interest in biochemical engineering and medical fields. This research is the result of an international collaboration with Professor Madoka Takai of the University of Tokyo, Professor Dirk Linke of the University of Oslo (Norway), Professor Stephan Göttig and Professor Volkhard A. J. Kempf of Goethe University (Germany).
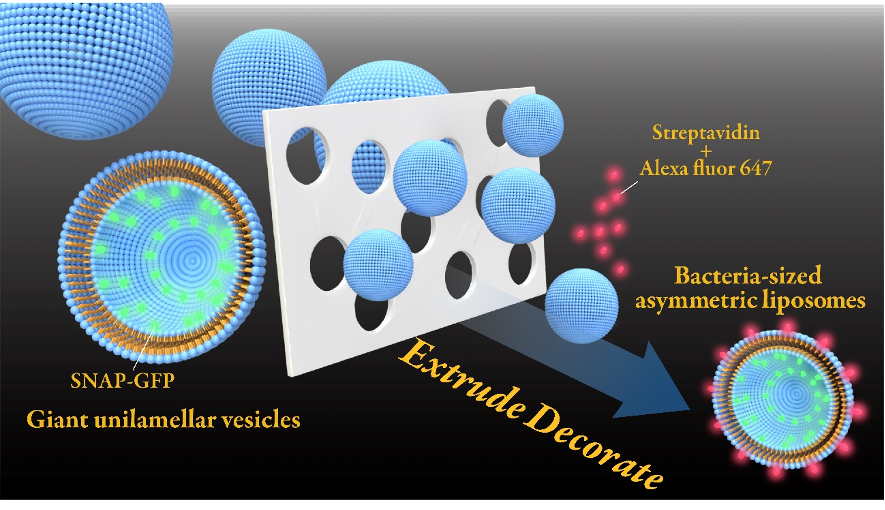
- In artificial cell creation, liposomes, consisting of a single lipid membrane with a diameter of over 10 μm, have typically been used. However, there have been technical constraints in creating artificial cells that mimic the size and membrane structure of bacterial cells, which are about 1 μm in diameter. In this study, by combining the interface transfer method, a universal technique for liposome production, with the extruder method, we developed a simple method to produce asymmetric liposomes with different proteins localized on the inner and outer layers of the lipid bilayer. This research represents a step towards the creation of artificial bacterial cells and is expected to contribute to the analysis of bacterial membrane structures and the functions of proteins therein. This research is a collaborative effort with the Matsuura Laboratory at Tokyo Institute of Technology and the Tanaka Laboratory at Tohoku University.
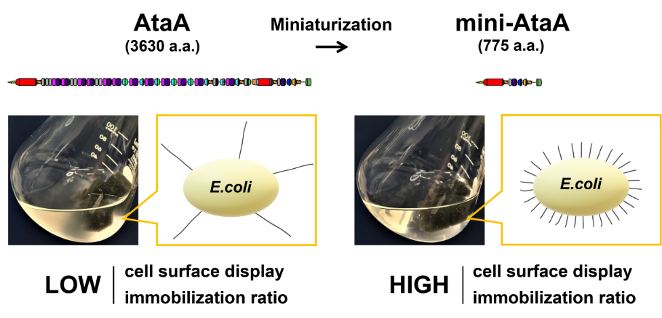
- In our laboratory, we have developed a simple, powerful, and reversible method for microbial immobilization by growing a unique nanofiber protein, AtaA, on bacterial cells. However, due to AtaA's large size, formed by a poly-peptide chain of 3630 amino acid residues that assemble into a homotrimer, it could only be grown on a limited range of bacteria. In this study, we successfully identified the adhesive sites of AtaA and downsized it to 775 amino acid residues while retaining its function. This enabled us to grow the downsized AtaA in industrially useful bacteria such as E. coli without decreasing their growth rate or other enzyme activities, making it applicable for immobilized microbial reactions. This research is expected to accelerate the creation of environmentally friendly bioprocesses and contribute to achieving carbon neutrality. This study is an international collaboration with Professor Andrei N. Lupas from the Max Planck Institute in Germany and Professor Dirk Linke from the University of Oslo in Norway.
- Acinetobacter sp. Tol 5, discovered in the Hori lab, has unique properties that allow it to adhere to various material surfaces via the nanofiber protein AtaA. However, because Tol 5 is also self-agglomerating, it has not been possible to evaluate the pure interaction between Tol 5 and the surface of materials using the conventional adhesion test method, which measures the number of bacteria attached to the surface. In this study, the adhesion force between Tol 5 and the material surface was measured at the single-cell level in liquid using an atomic force microscope (AFM), which can measure nanometer-sized irregularities and picoNewton-level forces scanning the sample surface with a fine tip. As a result, it was found that Tol 5 exhibited significantly stronger adhesion than other bacteria. Furthermore, it was found that the low ionic strength environment and the casamino acid solution, which were known to reduce the adhesion of Tol 5, acted through different mechanisms. As a result of this study, a better understanding of AtaA-mediated adhesion of Tol 5 is expected to develop AtaA-based bacterial immobilization technology further.
- We have sequenced the complete genome of the highly adhesive bacterium Acinetobacter sp. Tol 5, which was discovered in the Hori lab. Because the genome of Tol 5 contains an extensive repeat sequence, next-generation sequencers (NGS), which can only analyze short reads, have not been able to determine the full-length genome. In this study, we determined the full-length genome with high accuracy by using the MinION nanopore sequencer, which can analyze long reads, and the iSeq 100, the top model NGS in the lab. In the past, outsourced analysis was done genome sequencing, but the Hori laboratory, equipped with MinION and iSeq 100, can determine the genome within a few days. The defined genome sequence is expected to advance the research on Tol 5 further.
- The gas-phase microbial reaction is an innovative bioprocess developed in the Hori laboratory. By removing the liquid phase from the reactor, the diffusion rate of gaseous molecules can be increased, dramatically improving the performance of the microbial reaction. Under such reaction conditions, microorganisms are expected to have different metabolic kinetics from conventional microbial reaction systems (i.e., those with a liquid phase). In this study, we used metabolomic analysis of Methylococcus capsulatus (Bath), a representative methanogen, to clarify the changes in metabolic dynamics during gas-phase microbial reactions. We found that the metabolic state of the gas-phase reaction was significantly different from that of the liquid-phase reaction, suggesting that the metabolic state was more suitable for material production. The results of this study provide hints for further improving the efficiency of gas-phase microbial reactions and the production of new substances. This research was conducted in collaboration with Bamba Laboratory, Kyushu University.
- Genetically engineered microorganisms (GEMs) are strictly restricted from leaking into the environment due to the risk of disturbing the ecosystem. Therefore, although many valuable GEMs have been developed through genetic modification technology, we have not used GEMs for environmental cleanup. Biological containment" is a technology that makes GEMs live only in a specific environment so that they do not affect the ecosystem. We thought that if we could create GEMs that degrade pollutants but can only live in the environment where the contaminants are present, we could use GEMs for environmental conservation (GEMs die after degrading pollutants). We developed a GEM that would not increase after degrading toluene in this research. The GEM we set significantly limited the growth of toluene after it was contaminated, but we could not suppress it entirely. To find the cause of this, we conducted a genetic analysis using the nanopore sequencer MinION and discovered and proposed improvements. The results obtained are essential for developing biological containment technology for environmental protection.
S. Inoue, S. Yoshimoto, K. Hori
A new target of multiple lysine methylation in bacteria
J. Bacteriol. 207, e00325-24 (2025).
S. Yoshimoto, S. Ishii, A. Kawashiri, T. Matsushita, D. Linke, S. Göttig, VAJ. Kempf, M. Takai and K. Hori
Adhesion preference of the sticky bacterium Acinetobacter sp. Tol 5
Front. Bioeng. Biotechnol. 12, 1342418 (2024).
K. Noba, S. Yoshimoto, Y. Tanaka, T. Yokoyama, T. Matsuura, and K. Hori
Simple Method for the Creation of a Bacteria-Sized Unilamellar Liposome with Different Proteins Localized to the Respective Sides of the Membrane
ACS Synth. Biol. 12, (2023) 1437-1446
S. Yoshimoto, S. Aoki, Y. Ohara, M. Ishikawa, A. Suzuki, D. Linke, A. Lupas, and K. Hori
Identification of the adhesive domain of AtaA from Acinetobacter sp. Tol 5 and its application in immobilizing Escherichia coli
Front. Bioeng. Biotechnol. 10, 1095057 (2023)
S. Ishii, S. Yoshimoto and K. Hori
Single-cell adhesion force mapping of a highly sticky bacterium in liquid
J Colloid Interface Sci., 606, (2022) 628-634

M. Ishikawa and K. Hori
Complete Genome Sequence of the Highly Adhesive Bacterium Acinetobacter sp. Strain Tol 5
Microbiol Resour Announc, 10, (2021) e00567-21
Y.-Y. Chen, Y. Soma, M. Ishikawa, M. Takahashi, Y. Izumi, T. Bamba and K. Hori
Metabolic alteration of Methylococcus capsulatus str. Bath during a microbial gas-phase reaction
Biores. Technol., 330, (2021) 125002
M. Ishikawa, T. Kojima, and K. Hori
Development of a Biocontained Toluene-Degrading Bacterium for Environmental Protection
Microbiol Spectr., 9, (2021) e00259-21
